Predicting the efficacy of trivalent influenza vaccines
- Area
- Systems Pharmacology
- Expertise
- Biomarker identification
Challenge
Extract a transcriptional biomarker to predict the efficacy of trivalent influenza vaccines and expand our understanding of the early events following the vaccination.
What we did
By applying our normalization-free algorithm for biomarker identification to a transcriptomic dataset from peripheral blood mononuclear cells (PBMCs), we identified a biomarker whose early response was able to discriminate low- from high-responder subjects one month after the vaccination. The identified biomarker was then validated using a separate study including vaccination campaigns spanning a total of three years.
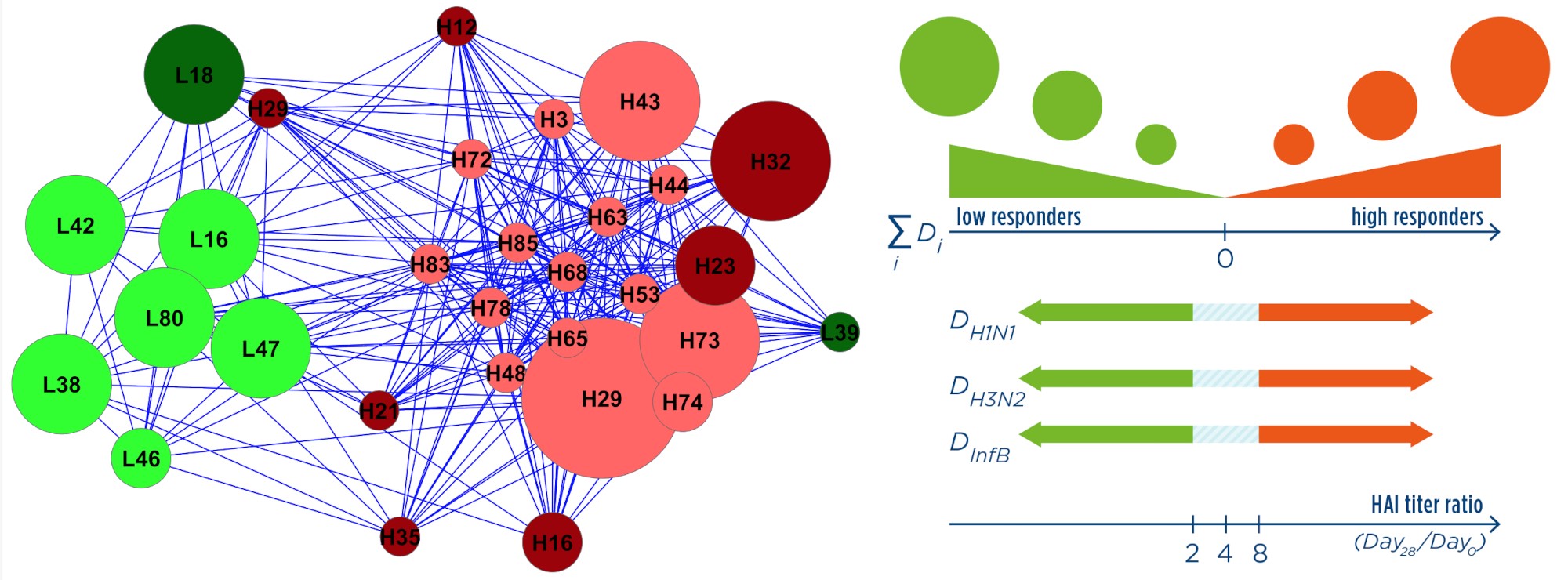
Outcome
From the analysis of the biomarker, we identified pre-existing immunity against one or more of the vaccine antigens as a confounding factor. We also introduced a composite response measure, which better accounts for the responses to the different strains represented in the vaccine with respect to other indices introduced in literature.
Partner

GSK is a science-led global healthcare company with a special purpose to improve the quality of human life by helping people do more, feel better, live longer.
https://it.gsk.com/it-it/Publications
Marchetti L, Siena E, Lauria M, Maffione D, Pacchiani N, Priami C, Medini D, Exploring the Limitations of Peripheral Blood Transcriptional Biomarkers in Predicting Influenza Vaccine Responsiveness, Complexity, vol. 2017, Article ID 3017632, 9 pages, 2017. https://doi.org/10.1155/2017/3017632