Investigating Duchenne muscular dystrophy biology through plasma proteome
- Area
- Systems Pharmacology
- Expertise
- Biomarker identification
- Pathway analysis
Challenge
Identify disease biomarkers and deregulated pathways in Duchenne muscular dystrophy from omic data
What we did
We developed a two-step computational workflow to extract biological knowledge from proteome profiles. In the first step we applied SCUDO, our proprietary rank-based classification method, to identify disease biomarkers. In the second step, we ran NASFinder, a network-based functional pathway analysis algorithm we developed to analyze the connections among genes of interest and their upstream regulators. NASFinder allowed us to explore the interactions among the proteins in the biomarker panel and identify affected pathways and candidate transcription factors regulating the expression of the genes encoding the disease biomarkers.
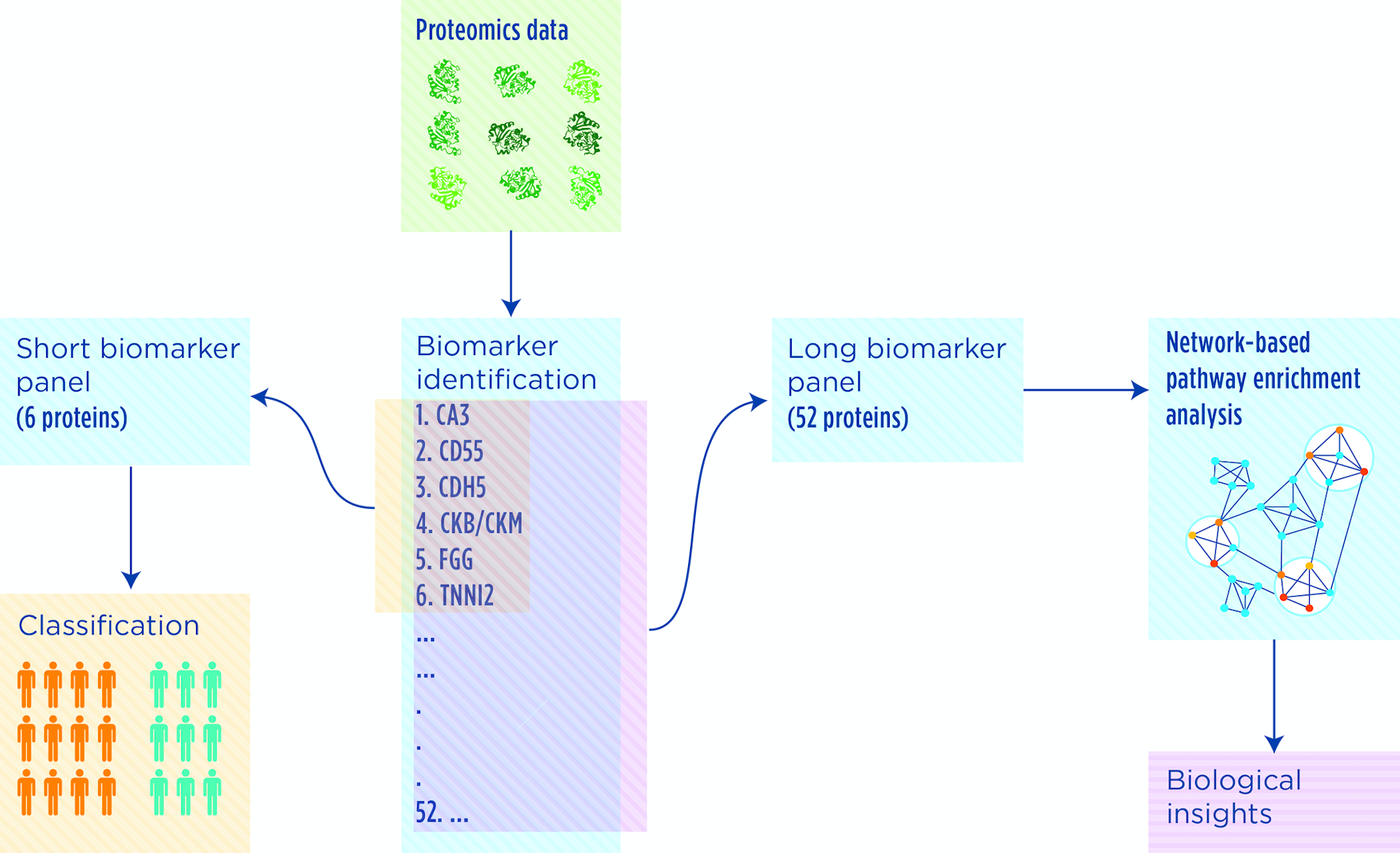
Outcome
The workflow was applied to a dataset of serum proteome profiles obtained from a cohort of Duchenne muscular dystrophy patients and controls. We identified a biomarker panel composed of six proteins, namely CA3, CD55, CDH5, CKB/CKM, FGG and TNNI2, able to distinguish patients from controls. In addition to muscle-related functions, NASFinder revealed other biological processes such as apoptosis, signaling in the immune system and neurotrophin signaling as significantly modulated in patients compared with controls and pointed out the transcription factor. FoxO as a candidate regulator of different pathways.
Publications
Parolo S, Marchetti L, Lauria M, Misselbeck K, Scott-Boyer MP, Caberlotto L, Priami C. Combined use of protein biomarkers and network analysis unveils deregulated regulatory circuits in Duchenne muscular dystrophy. PLoS One. 2018 Mar 12;13(3):e0194225. https://doi.org/10.1371/journal.pone.0194225.