Developing a novel unsupervised and normalization-free tool for multi-omics integration and biomarker identification
- Area
- Systems Pharmacology
- Expertise
- Biomarker identification
Challenge
Modern profiling technologies have led to relevant progress toward precision medicine and disease management. A new trend in patient classification is to integrate multiple data types for the same subjects to increase the chance of identifying meaningful phenotype groups.
What we did
We developed MOUSSE (Multi-Omics Using Subject-Specific SignaturEs), a new unsupervised and normalization-free tool for multi-omics integration able to maintain good clustering performance across a wide range of omics data. The main innovations are the use of rank-based subject-specific signatures and the use of such signatures to derive subject similarity networks and extract multi-omics biomarkers.
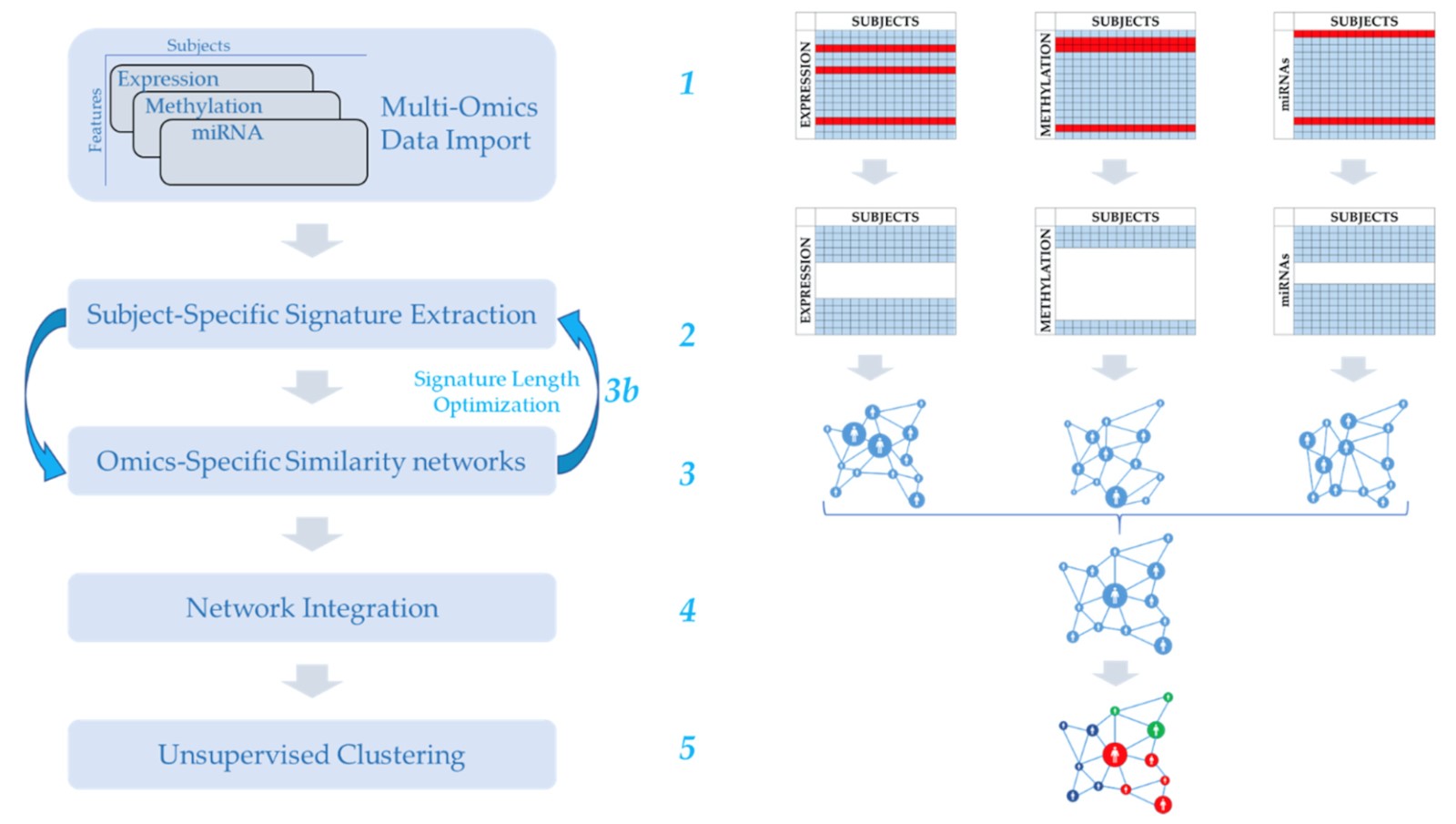
Outcome
We applied our method to analyze survival time in ten different types of cancer. We produced a meaningful clusterization of the subjects and obtained a higher average classification score than ten state-of-the-art algorithms tested on the same data. In addition, we extracted from the computed biomarkers a list of relevant biological features that were later identified as highly involved in cancer progression and differential survival.
Publications
Fiorentino G, Visintainer R, Domenici E, Lauria M, Marchetti L, MOUSSE: Multi-Omics Using Subject-Specific SignaturEs, Cancers 2021, 13, 3423. https://doi.org/10.3390/cancers13143423